pymcmcstat.samplers package¶
pymcmcstat.samplers.Adaptation module¶
Created on Thu Jan 18 11:14:11 2018
@author: prmiles
-
class
pymcmcstat.samplers.Adaptation.Adaptation[source]¶ Bases:
objectAdaptive Metropolis (AM) algorithm based on [HST+01].
- Attributes:
-
run_adaptation(covariance, options, isimu, iiadapt, rejected, chain, chainind, u, npar, alpha)[source]¶ Run adaptation step
- Args:
- covariance (
CovarianceProcedures): Covariance methods and variables - options (
SimulationOptions): Options for MCMC simulation - isimu (
int): Simulation counter - iiadapt (
int): Adaptation counter - rejected (
dict): Rejection counter - chain (
ndarray): Sampling chain - chainind (
ind): Relative point in chain - u (
ndarray): Latest random sample points - npar (
int): Number of parameters being sampled - alpha (
float): Latest Likelihood evaluation
- covariance (
- Returns:
- covariance (
CovarianceProcedures): Updated covariance object
- covariance (
-
pymcmcstat.samplers.Adaptation.adjust_cov_matrix(upcov, R, npar, qcov_adjust, qcov_scale, rejected, iiadapt, verbosity)[source]¶ Adjust covariance matrix if found to be singular.
- Args:
- upcov (
ndarray): Parameter covariance matrix. - R (
ndarray): Cholesky decomposition of covariance matrix. - npar (
int): Number of parameters. - qcov_adjust (
float): Covariance adjustment factor. - qcov_scale (
float): Scale parameter - rejected (
dict): Rejection counters. - iiadapt (
int): Adaptation counter. - verbosity (
int): Verbosity of display output.
- upcov (
- Returns:
- R (
ndarray): Cholesky decomposition of covariance matrix.
- R (
-
pymcmcstat.samplers.Adaptation.below_burnin_threshold(rejected, iiadapt, R, burnin_scale, verbosity)[source]¶ Update Cholesky Matrix using below burnin thershold
-
pymcmcstat.samplers.Adaptation.check_for_singular_cov_matrix(upcov, R, npar, qcov_adjust, qcov_scale, rejected, iiadapt, verbosity)[source]¶ Check if singular covariance matrix
- Args:
- upcov (
ndarray): Parameter covariance matrix. - R (
ndarray): Cholesky decomposition of covariance matrix. - npar (
int): Number of parameters. - qcov_adjust (
float): Covariance adjustment factor. - qcov_scale (
float): Scale parameter - rejected (
dict): Rejection counters. - iiadapt (
int): Adaptation counter. - verbosity (
int): Verbosity of display output.
- upcov (
- Returns:
- R (
ndarray): Adjusted Cholesky decomposition of covariance matrix.
- R (
-
pymcmcstat.samplers.Adaptation.initialize_covariance_mean_sum(x, w)[source]¶ Initialize covariance chain, local mean, local sum
-
pymcmcstat.samplers.Adaptation.is_semi_pos_def_chol(x)[source]¶ Check if matrix is semi-positive definite using Cholesky Decomposition
-
pymcmcstat.samplers.Adaptation.scale_cholesky_decomposition(Ra, qcov_scale)[source]¶ Scale Cholesky decomposition
-
pymcmcstat.samplers.Adaptation.setup_cholupdate(R, oldwsum, wsum, oldmean, xi)[source]¶ Setup input arguments to the Cholesky update function
- Args:
- Returns:
-
pymcmcstat.samplers.Adaptation.setup_w_R(w, oldR, n)[source]¶ Setup weights and Cholesky matrix
- Args:
- Returns:
-
pymcmcstat.samplers.Adaptation.unpack_covariance_settings(covariance)[source]¶ Unpack covariance settings
- Args:
- covariance (
CovarianceProcedures): Covariance methods and variables
- covariance (
- Returns:
- last_index_since_adaptation (
int): Last index since adaptation occured. - R (
ndarray): Cholesky decomposition of covariance matrix. - oldcovchain (
ndarray): Covariance matrix history. - oldmeanchain (
ndarray): Current mean chain values. - oldwsum (
ndarray): Weights - no_adapt_index (
numpy.ndarray): Indices of parameters not being adapted. - qcov_scale (
float): Scale parameter - qcov (
ndarray): Covariance matrix
- last_index_since_adaptation (
-
pymcmcstat.samplers.Adaptation.unpack_simulation_options(options)[source]¶ Unpack simulation options
- Args:
- options (
SimulationOptions): Options for MCMC simulation
- options (
- Returns:
- burnintime (
int): - burnin_scale (
float): Scale for burnin. - ntry (
int): Number of tries to take before rejection. Default is method dependent. - drscale (
ndarray): Reduced scale for sampling in DR algorithm. Default is [5,4,3]. - alphatarget (
float): Acceptance ratio target. - etaparam (
float): - qcov_adjust (
float): Adjustment scale for covariance matrix. - doram (
int): Flag to perform'ram'algorithm (Obsolete). - verbosity (
int): Verbosity of display output.
- burnintime (
-
pymcmcstat.samplers.Adaptation.update_cov_from_covchain(covchain, qcov, no_adapt_index)[source]¶ Update covariance matrix from covariance matrix chain
- Args:
- covchain (
ndarray): Covariance matrix history. - qcov (
ndarray): Parameter covariance matrix - no_adapt_index (
numpy.ndarray): Indices of parameters not being adapted.
- covchain (
- Returns:
- upcov (
ndarray): Updated covariance matrix
- upcov (
-
pymcmcstat.samplers.Adaptation.update_cov_via_ram(u, isimu, etaparam, npar, alphatarget, alpha, R)[source]¶ Update covariance matrix via RAM
- Args:
- Returns:
- upcov (
ndarray): Updated parameter covariance matrix.
- upcov (
-
pymcmcstat.samplers.Adaptation.update_covariance_mean_sum(x, w, oldcov, oldmean, oldwsum, oldR=None)[source]¶ Update covariance chain, local mean, local sum
- Args:
- Returns:
pymcmcstat.samplers.DelayedRejection module¶
Created on Thu Jan 18 10:42:07 2018
@author: prmiles
-
class
pymcmcstat.samplers.DelayedRejection.DelayedRejection[source]¶ Bases:
objectDelayed Rejection (DR) algorithm based on [HLMS06].
-
classmethod
initialize_next_metropolis_step(npar, old_theta, sigma2, RDR)[source]¶ Take metropolis step according to DR
- Args:
- Returns:
- next_set (
ParameterSet): New proposal set - u (
numpy.ndarray): Numbers sampled from standard normal distributions (u.shape = (1,npar))
- next_set (
-
run_delayed_rejection(old_set, new_set, RDR, ntry, parameters, invR, sosobj, priorobj, custom=None)[source]¶ Perform delayed rejection step - occurs in standard metropolis is not accepted.
- Args:
- old_set (
ParameterSet): Features of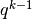
- new_set (
ParameterSet): Features of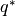
- RDR (
ndarray): Cholesky decomposition of parameter covariance matrix for DR steps - ntry (
int): Number of DR steps to perform until rejection - parameters (
ModelParameters): Model parameters - invR (
ndarray): Inverse Cholesky decomposition matrix - sosobj (
SumOfSquares): Sum-of-Squares function - priorobj (
PriorFunction): Prior function
- old_set (
- Returns:
- accept (
int): 0 - reject, 1 - accept - out_set (
ParameterSet): If accept == 1, then latest DR set; Else,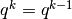
- outbound (
int): 1 - rejected due to sampling outside of parameter bounds
- accept (
-
classmethod
-
pymcmcstat.samplers.DelayedRejection.extract_state_elements(iq, stage, trypath)[source]¶ Extract elements from tried paths.
-
pymcmcstat.samplers.DelayedRejection.log_posterior_ratio(x1, x2)[source]¶ Calculate the logarithm of the posterior ratio.
- Args:
- x1 (
ParameterSet): Old set -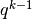
- x2 (
ParameterSet): New set -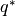
- x1 (
- Returns:
- zq (
float): Logarithm of posterior ratio.
- zq (
-
pymcmcstat.samplers.DelayedRejection.nth_stage_log_proposal_ratio(iq, trypath, invR)[source]¶ Gaussian nth stage log proposal ratio.
Logarithm of
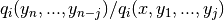
-
pymcmcstat.samplers.DelayedRejection.update_set_based_on_acceptance(accept, old_set, next_set)[source]¶ Define output set based on acceptance
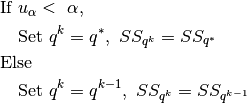
- Args:
- accept (
int): 0 - reject, 1 - accept - old_set (
ParameterSet): Features of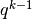
- next_set (
ParameterSet): New proposal set
- accept (
- Returns:
- out_set (
ParameterSet): If accept == 1, then latest DR set; Else,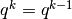
- out_set (
pymcmcstat.samplers.Metropolis module¶
Created on Thu Jan 18 10:30:29 2018
@author: prmiles
-
class
pymcmcstat.samplers.Metropolis.Metropolis[source]¶ Bases:
objectPseudo-Algorithm:
- Sample
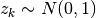
- Construct candidate
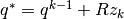
- Compute
![\quad SS_{q^*} = \sum_{i=1}^N[v_i-f_i(q^*)]^2](_images/math/b1c1f5436263be391adfda47c68cec9bf4849fc4.png)
- Compute
![\quad \alpha = \min\Big(1, e^{[SS_{q^*} - SS_{q^{k-1}}]/(2\sigma^2_{k-1})}\Big)](_images/math/714efef0b699780299395351aec662a1252b8c4e.png)
- If
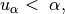
- Set
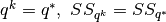
- Else
- Set
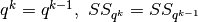
- If
- Attributes:
acceptance_test()run_metropolis_step()unpack_set()
-
classmethod
calculate_posterior_ratio(ss1, ss2, sigma2, newprior, oldprior)[source]¶ Calculate acceptance ratio
![\alpha = \min\Big[1, \frac{\mathcal{L}(\nu_{obs}|q^*, \sigma_{k-1}^2)\pi_0(q^*)}{\mathcal{L}(\nu_{obs}|q^{k-1}, \sigma_{k-1}^2)\pi_0(q^{k-1})}\Big]](_images/math/02297ac3399a08bbc24d6284133dcc6467dee9ba.png)
where the Gaussian likelihood function is
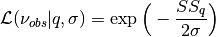
and Gaussian prior function is
![\pi_0(q) = \exp \Big[-\frac{1}{2}\Big(\frac{q - \mu_0}{\sigma_0}\Big)^2\Big].](_images/math/1144b68429d43af73a204d13d803ac5a0fa49230.png)
For the Gaussian likelihood and prior, this yields the acceptance ratio
![\alpha = \exp\Big[-0.5\Big(\sum\Big(\frac{ SS_{q^*} - SS_{q^{k-1}} }{ \sigma_{k-1}^2 }\Big) + p_1 - p_2\Big)\Big].](_images/math/73df299f0339424ad0c1b6e90c98c5a9f5aeb21b.png)
For more details regarding the prior function, please refer to the
PriorFunctionclass.Note
The default behavior of the package is to use Gaussian likelihood and prior functions (as of v1.8.0). Future releases will expand the functionality to allow for alternative likelihood and prior definitions.
- Args:
- Returns:
- alpha (
float): Result of likelihood function
- alpha (
-
run_metropolis_step(old_set, parameters, R, prior_object, sos_object, custom=None)[source]¶ Run Metropolis step.
- Args:
- old_set (
ParameterSet): Features of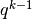
- parameters (
ModelParameters): Model parameters - R (
ndarray): Cholesky decomposition of parameter covariance matrix - priorobj (
PriorFunction): Prior function - sosobj (
SumOfSquares): Sum-of-Squares function
- old_set (
- Returns:
- accept (
int): 0 - reject, 1 - accept - newset (
ParameterSet): Features of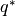
- outbound (
int): 1 - rejected due to sampling outside of parameter bounds - npar_sample_from_normal (
ndarray): Latet random sample points
- accept (
- Sample
pymcmcstat.samplers.SamplingMethods module¶
Created on Thu Jan 18 10:11:40 2018
@author: prmiles
pymcmcstat.samplers.utilities module¶
Created on Wed Jun 27 12:07:11 2018
Utility functions used by different samplers
@author: prmiles
-
pymcmcstat.samplers.utilities.calculate_log_posterior_ratio(loglikestar, loglike, logpriorstar, logprior)[source]¶ Calculate log posterior ratio:
![\log(\alpha) = \min\Big[0, \log(\mathcal{L}(\nu_{obs}|q^*)) + \log(\pi_0(q^*)) - \log(\mathcal{L}(\nu_{obs}|q^{k-1})) - \log(\pi_0(q^{k-1}))\Big]](_images/math/8164a881779be15f452c5e104055821ddd0b867b.png)
For more details regarding the prior and likelihood functions, please refer to the
PriorFunctionandLikelihoodFunctionclass, respectively.Note
The default behavior of the package is to use Gaussian likelihood and prior functions (as of v1.8.0). Future releases will expand the functionality to allow for alternative likelihood and prior definitions.
-
pymcmcstat.samplers.utilities.is_sample_outside_bounds(theta, lower_limits, upper_limits)[source]¶ Check whether proposal value is outside parameter limits
-
pymcmcstat.samplers.utilities.log_posterior_ratio_acceptance_test(alpha)[source]¶ Run log posterior ratio acceptance test
-
pymcmcstat.samplers.utilities.posterior_ratio_acceptance_test(alpha)[source]¶ Run posterior ratio acceptance test
-
pymcmcstat.samplers.utilities.sample_candidate_from_gaussian_proposal(npar, oldpar, R)[source]¶ Sample candidate from Gaussian proposal distribution
-
pymcmcstat.samplers.utilities.set_outside_bounds(next_set)[source]¶ Assign set features based on being outside bounds
- Args:
- next_set (
ParameterSet):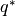
- next_set (
- Returns:
- next_set (
ParameterSet):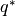 with updated features
with updated features - outbound (
bool): True
- next_set (
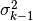
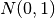 .
.